News
Neural Transformation Models (one 🔨 for many nails )
A valuable way to describe a probability distribution is by considering it as a transformation from a basic distribution to a complex distribution. Collaborating with Beate Sick and others, we have employed networks to determine the parameters θ of a transformation function since 2020 and have found it to be a versatile tool for many applications.
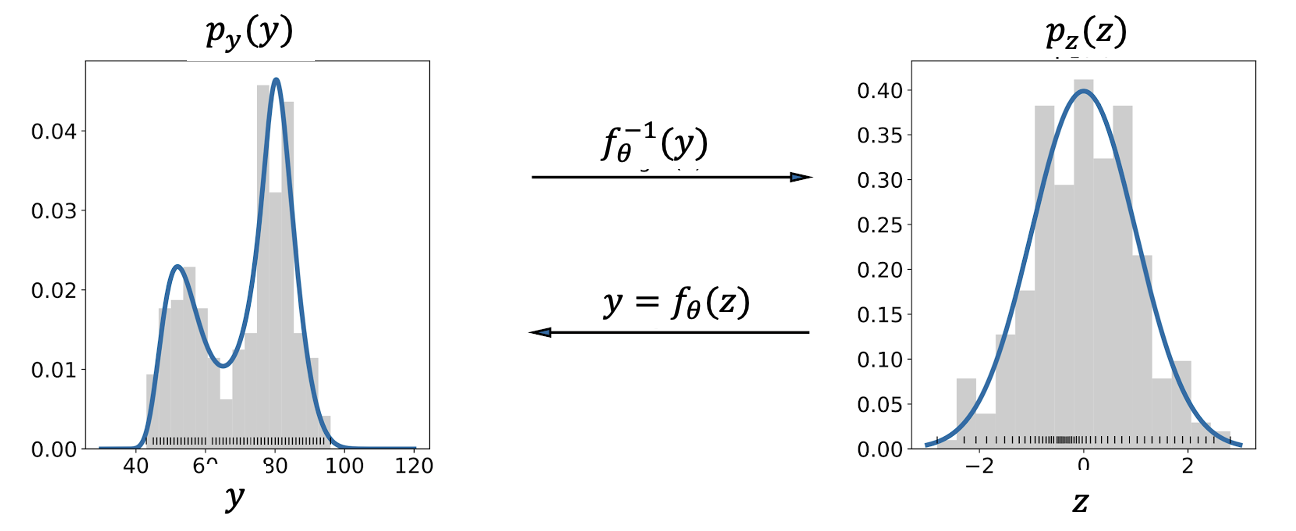
- To create a flexible output distribution for a neural network (NN):
- We introduced this concept in using it to predict a person’s age based on an image.
- To model the power consumption forecasted by a neural network
- For comprehensible integration of NN and interpretable coefficients:
-
To model flexible distributions for variational inference
- To model the functions of a structural causal model in an interpretable way TramDAM
Book on probabilistic deep learning
Our Book on probabilistic deep learning is finally out, it describes how to combine probabilistic modelling with deep learning.

https://www.manning.com/books/probabilistic-deep-learning-with-python
You can find the Jupyter notebooks here
Assigning uncertainty in deep learning
Recently deep neural networks have become standard in many areas of analysing perception data like images or audio. However, they can be spectacular wrong.
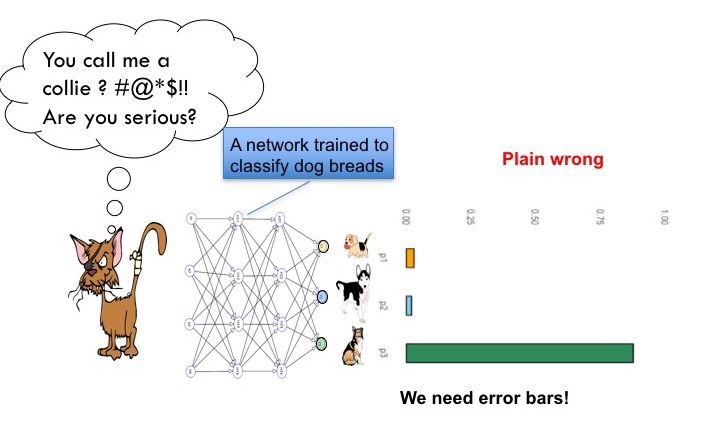 The network overconfidently assigns a high probability to the wrong class. I get scared when I think of self driving cars being overconfident. While standard networks cannot state their uncertainty of a prediction, novel methods allow to include uncertainty information. One way to do so is to use dropout also during test time. [Yarin Gal has shown that this allows to quantify uncertainty in his Phd thesis](https://arxiv.org/abs/1506.02142).
The network overconfidently assigns a high probability to the wrong class. I get scared when I think of self driving cars being overconfident. While standard networks cannot state their uncertainty of a prediction, novel methods allow to include uncertainty information. One way to do so is to use dropout also during test time. [Yarin Gal has shown that this allows to quantify uncertainty in his Phd thesis](https://arxiv.org/abs/1506.02142).
We use this approach in several places:
-
Paper 2018 for the use in high content screening: “Know When You Don’t Know: A Robust Deep Learning Approach in the Presence of Unknown Phenotypes” ASSAY and Drug Development Technologies.
-
Poster 2018 (Swiss Data Science Conference, Laussane) “Are you serious”
-
Invited talk 2017 “Deep learning for single cell phenotype classification in High-Content Screening” at SIBS 2017
And in other ongoing projects, stay tuned.
Deep Learning for HCS
In high content phenotypic screening millions of cells need to be classified based on their morphology. Images of the cells are taken at different 5 ‘color channels’ using different fluorescent markers.
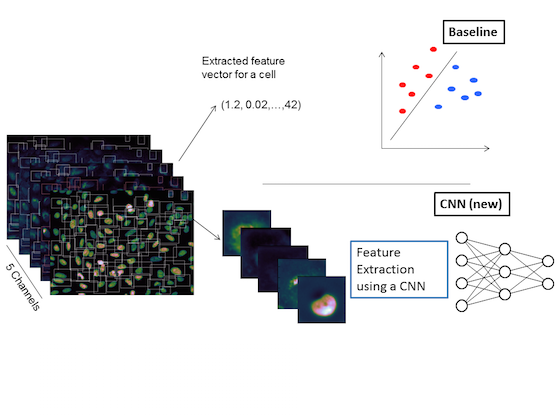
In this project we compared the conventional pipeline (upper branch) against a deep learning approach (lower branch). For the deep learning approach the construction of appropriate feature definitions is part of the training. Whereas, in the traditional pipeline expert knowledge is required for the tedious creation of handcrafted features. Compared to the best traditional method, the misclassification rate in the deep learning approach is reduced from 8.9% to 6.6%.
For more information: See our Poster and talk at the SIBS 2015 in Basel or have a look at out paper: Dürr, O., and Sick, B. Single-cell phenotype classification using deep convolutional neural networks. Journal of Biomolecular Screening (2016).
Pi-Vision (Deep Learning on a Raspberry Pi)
The pi-Vision project was my first research project applying deep learning. The task was to use a raspberry pi minicomputer for face recognition. We used among other approaches a convolution neural network, a recently developed technique revolutionising image recognition. The following video (please turn off the sound!) shows the trained network doing predictions.
For more information see: Dürr, Oliver, et al. “Deep Learning on a Raspberry Pi for Real Time Face Recognition.” Eurographics (Posters), 2015.
Using Community Structure for Complex Network Layout
We present a new layout algorithm for complex networks that combines a multi-scale approach for community detection with a standard force-directed design. Since community detection is computationally cheap, we can exploit the multi-scale approach to generate network configurations with close-to-minimal energy very fast. As a further asset, we can use the knowledge of the community structure to facilitate the interpretation of large networks, for example the network defined by protein-protein interactions.
Below you find a video showing the algorithm at work, disentangling the network of streets in the UK, having 4824 vertices and 6827 nodes.
For more information see: Dürr, Oliver, and Arnd Brandenburg. “Using Community Structure for Complex Network Layout.” arXiv preprint arXiv:1207.6282 (2012).